Reading data files
Opening files
You will normally access data from a run, which is stored as a directory
containing HDF5 files. You can open a run using RunDirectory() with the
path of the directory, or using open_run() with the proposal number and
run number to look up the standard data paths on the Maxwell cluster.
- extra_data.RunDirectory(path, include='*', file_filter=<function lc_any>, *, inc_suspect_trains=True, parallelize=True, _use_voview=True)
Open data files from a ‘run’ at European XFEL.
run = RunDirectory("/gpfs/exfel/exp/XMPL/201750/p700000/raw/r0001")
A ‘run’ is a directory containing a number of HDF5 files with data from the same time period.
Returns a
DataCollectionobject.- Parameters:
path (str) – Path to the run directory containing HDF5 files.
include (str) – Wildcard string to filter data files.
file_filter (callable) – Function to subset the list of filenames to open. Meant to be used with functions in the extra_data.locality module.
inc_suspect_trains (bool) – If False, suspect train IDs within a file are skipped. In newer files, trains where INDEX/flag are 0 are suspect. For older files which don’t have this flag, out-of-sequence train IDs are suspect. If True (default), it tries to include these trains.
parallelize (bool) – Enable or disable opening files in parallel. Particularly useful if creating child processes is not allowed (e.g. in a daemonized
multiprocessing.Process).
- extra_data.open_run(proposal, run, data='raw', include='*', file_filter=<function lc_any>, *, inc_suspect_trains=True, parallelize=True, aliases='{}/usr/extra-data-aliases.yml', _use_voview=True)
Access EuXFEL data on the Maxwell cluster by proposal and run number.
run = open_run(proposal=700000, run=1)
Returns a
DataCollectionobject.- Parameters:
proposal (str, int) – A proposal number, such as 2012, ‘2012’, ‘p002012’, or a path such as ‘/gpfs/exfel/exp/SPB/201701/p002012’.
run (str, int) – A run number such as 243, ‘243’ or ‘r0243’.
data (str or Sequence of str) – ‘raw’, ‘proc’ (processed), or any other location relative to the proposal path with data per run to access. May also be ‘all’ (both ‘raw’ and ‘proc’) or a sequence of strings to load data from several locations, with later locations overwriting sources present in earlier ones.
include (str) – Wildcard string to filter data files.
file_filter (callable) – Function to subset the list of filenames to open. Meant to be used with functions in the extra_data.locality module.
inc_suspect_trains (bool) – If False, suspect train IDs within a file are skipped. In newer files, trains where INDEX/flag are 0 are suspect. For older files which don’t have this flag, out-of-sequence train IDs are suspect. If True (default), it tries to include these trains.
parallelize (bool) – Enable or disable opening files in parallel. Particularly useful if creating child processes is not allowed (e.g. in a daemonized
multiprocessing.Process).aliases (str, Path) – Path to an alias file for the run, see the documentation for
DataCollection.with_aliases()for details. If the argument is a string with a format argument like{}/path/to/aliases.yml, then the format argument will be replaced with the proposal directory path. By default it looks for a file named{}/usr/extra-data-aliases.yml.
You can also open a single file. The methods described below all work for either a run or a single file.
- extra_data.H5File(path, *, inc_suspect_trains=True)
Open a single HDF5 file generated at European XFEL.
file = H5File("RAW-R0017-DA01-S00000.h5")
Returns a
DataCollectionobject.- Parameters:
path (str) – Path to the HDF5 file
inc_suspect_trains (bool) – If False, suspect train IDs within a file are skipped. In newer files, trains where INDEX/flag are 0 are suspect. For older files which don’t have this flag, out-of-sequence train IDs are suspect. If True (default), it tries to include these trains.
See ‘Suspect’ train IDs for more details about the inc_suspect_trains
parameter.
Data structure
A run (or file) contains data from various sources, each of which has keys.
For instance, SA1_XTD2_XGM/XGM/DOOCS is one source, for an ‘XGM’ device
which monitors the beam, and its keys include beamPosition.ixPos and
beamPosition.iyPos.
European XFEL produces ten pulse trains per second, each of which can contain up to 2700 X-ray pulses. Each pulse train has a unique train ID, which is used to refer to all data associated with that 0.1 second window.
- class extra_data.DataCollection
- train_ids
A list of the train IDs included in this data. The data recorded may not be the same for each train.
- control_sources
A set of the control source names in this data, in the format
"SA3_XTD10_VAC/TSENS/S30100K". Control data is always recorded exactly once per train.
- instrument_sources
A set of the instrument source names in this data, in the format
"FXE_DET_LPD1M-1/DET/15CH0:xtdf". Instrument data may be recorded zero to many times per train.
- all_sources
A set of names for both instrument and control sources. This is the union of the two sets above.
- keys_for_source(source)
Get a set of key names for the given source
If you have used
select()to filter keys, only selected keys are returned.Only one file is used to find the keys. Within a run, all files should have the same keys for a given source, but if you use
union()to combine two runs where the source was configured differently, the result can be unpredictable.
- get_data_counts(source, key)
Get a count of data points in each train for the given data field.
Returns a pandas series with an index of train IDs.
- train_timestamps(labelled=False)
Get approximate timestamps for each train
Timestamps are stored and returned in UTC (not local time). Older files (before format version 1.0) do not have timestamp data, and the returned data in those cases will have the special value NaT (Not a Time).
If labelled is True, they are returned in a pandas series, labelled with train IDs. If False (default), they are returned in a NumPy array of the same length as data.train_ids.
- run_metadata() dict
Get a dictionary of metadata about the run
From file format version 1.0, the files capture: creationDate, daqLibrary, dataFormatVersion, karaboFramework, proposalNumber, runNumber, sequenceNumber, updateDate.
New in version 1.6.
- info(details_for_sources=())
Show information about the selected data.
- alias
Enables item access via source and key aliases.
- with_aliases(*alias_defs)
Apply aliases for convenient source and key access.
Allows to define aliases for sources or source-key combinations that may be used instead of their literal names to retrieve
SourceDataandKeyDataobjects viaDataCollection.alias.Multiple alias definitions may be passed as positional arguments in different formats:
Passing a dictionary mapping aliases to sources (passed as strings) or source-key pairs (passed as a 2-len tuple of strings).
Passing a string or PathLike pointing to a JSON, YAML (requires pyYAML installed) or TOML (requires Python 3.11 or with tomli installed) file containing the aliases. For unsupported formats, an
ImportErroris raised.The file should contain mappings from alias to sources as strings or source-key pairs as lists. In addition, source-key aliases may be defined by nested key-value pairs according to the respective format, shown here in YAML:
# Source alias. sa1-xgm: SA1_XTD2_XGM/XGM/DOOCS # Direct source key alias. sa1-intensity: [SA1_XTD2_XGM/XGM/DOOCS:output, data.intensityTD] # Nested source key alias, useful if you want aliases for multiple # keys of the same source. SA3_XTD10_MONO/MDL/PHOTON_ENERGY: mono-central-energy: actualEnergy
Returns a new
DataCollectionobject with the aliases for sources and keys.
- only_aliases(*alias_defs, strict=False, require_all=False)
Apply aliases and select only the aliased sources and keys.
A convenient function around
DataCollection.with_aliases()andDataCollection.select()applying both the aliases passed asalias_defsto the former and then selecting down theDataCollectionto any sources and/or their keys for which aliases exist.By default and unlike
DataCollection.select(), any sources or keys present in the alias definitions but not the data itself are ignored. This can be changed via the optional argumentstrict.The optional
require_allargument restricts the trains to those for which all selected sources and keys have at least one data entry. By default, all trains remain selected.Returns a new
DataCollectionobject with only the aliased sources and keys.
- drop_aliases()
Return a new DataCollection without any aliases.
Using aliases
Because source and key names are often quite long and obtuse, it can be useful
to define human-meaningful aliases for them. You can use
DataCollection.with_aliases() to add aliases to an existing
DataCollection, but it is often easier to define a per-proposal
aliases file. By default open_run() will look for a file named
usr/extra-data-aliases.yml under the proposal directory and load the aliases
in it, so by storing aliases in that file they will be loaded automatically
whenever opening a run with open_run().
Warning
Make sure that the aliases file is writable by everyone! Otherwise only the person who creates the file will be able to edit it. You can set loose permissions on it with this shell command:
chmod 666 extra-data-aliases.yml
You can then access sources and keys by aliases through the
DataCollection.alias property. For example, if this was in our
extra-data-aliases.yml file:
xgm: SA2_XTD1_XGM/XGM/DOOCS
MID_DET_AGIPD1M-1/DET/3CH0:xtdf:
agipd3-data: image.data
agipd3-mask: image.mask
Then we would be able to run:
run.alias["xgm"] # SourceData of the XGM
run.alias["agipd3-data"] # KeyData for image.data
run.alias["agipd3-mask"] # KeyData for image.mask
Calling repr() on run.alias will show all of the loaded aliases, along
with whether or not the aliases are actually present in the run. In practice
that means you can enter run.alias in a Jupyter notebook cell and it
will print something like this (note the ✗ for an invalid alias):
Loaded aliases:
✗ xgm: SA42_XTD1_XGM/XGM/DOOCS
MID_DET_AGIPD1M-1/DET/3CH0:xtdf:
agipd3-data: image.data
agipd3-mask: image.mask
Aliases can be used for selections as well, see
DataCollection.AliasIndexer.select() for more details.
Getting data by source & key
Selecting a source in a run gives a SourceData object.
You can use this to find keys belonging to that source:
xgm = run['SPB_XTD9_XGM/DOOCS/MAIN']
xgm.keys() # List the available keys
beam_x = xgm['beamPosition.ixPos'] # Get a KeyData object
Selecting a single source & key in a run gives a KeyData object.
You can get the data from this in various forms with the methods described
below, e.g.:
xgm_intensity = run['SA1_XTD2_XGM/XGM/DOOCS:output', 'data.intensityTD'].xarray()
- class extra_data.KeyData
- dtype
The NumPy dtype for this data. This indicates whether it contains integers or floating point numbers, and how many bytes of memory each number needs.
- ndim
The number of dimensions the data has. All data has at least 1 dimension (time). A sequence of 2D images would have 3 dimensions.
- shape
The shape of this data as a tuple, like for a NumPy array.
Finding the shape may require getting index data from several files
- entry_shape
The shape of a single entry in the data, e.g. a single frame from a camera. This is equivalent to
key.shape[1:], but may be quicker than that.
- data_counts(labelled=True)
Get a count of data entries in each train.
If labelled is True, returns a pandas series with an index of train IDs. Otherwise, returns a NumPy array of counts to match
.train_ids.
- nbytes
The number of bytes this data would take up in memory.
- size_mb
The size of the data in memory in megabytes.
- size_gb
The size of the data in memory in gigabytes.
- ndarray(roi=(), out=None)
Load this data as a numpy array
roi may be a
numpy.s_[]expression to load e.g. only part of each image from a camera. If out is not given, a suitable array will be allocated.
- series()
Load this data as a pandas Series. Only for 1D data.
- xarray(extra_dims=None, roi=(), name=None)
Load this data as a labelled xarray array or dataset.
The first dimension is labelled with train IDs. Other dimensions may be named by passing a list of names to extra_dims.
For scalar datatypes, an xarray.DataArray is returned using either the supplied name or the concatenated source and key name if omitted.
If the data is stored in a structured datatype, an xarray.Dataset is returned with a variable for each field. The data of these variables will be non-contiguous in memory, use Dataset.copy(deep=true) to obtain a contiguous copy.
- Parameters:
extra_dims (list of str) – Name extra dimensions in the array. The first dimension is automatically called ‘train’. The default for extra dimensions is dim_0, dim_1, …
roi (numpy.s_[], slice, tuple of slices, or by_index) – The region of interest. This expression selects data in all dimensions apart from the first (trains) dimension. If the data holds a 1D array for each entry, roi=np.s_[:8] would get the first 8 values from every train. If the data is 2D or more at each entry, selection looks like roi=np.s_[:8, 5:10] .
name (str) – Name the array itself. The default is the source and key joined by a dot. Ignored for structured data when a dataset is returned.
See also
- xarray documentation
How to use the arrays returned by
DataCollection.get_array()- Reading data to analyse in memory
Examples using xarray & pandas with EuXFEL data
- dask_array(labelled=False)
Make a Dask array for this data.
Dask is a system for lazy parallel computation. This method doesn’t actually load the data, but gives you an array-like object which you can operate on. Dask loads the data and calculates results when you ask it to, e.g. by calling a
.compute()method. See the Dask documentation for more details.If your computation depends on reading lots of data, consider creating a dask.distributed.Client before calling this. If you don’t do this, Dask uses threads by default, which is not efficient for reading HDF5 files.
- Parameters:
labelled (bool) – If True, label the train IDs for the data, returning an xarray.DataArray object wrapping a Dask array.
See also
- Dask Array documentation
How to use the objects returned by
DataCollection.get_dask_array()- Averaging detector data with Dask
An example using Dask with EuXFEL data
- train_id_coordinates()
Make an array of train IDs to use alongside data from
.ndarray().train_idsincludes each selected train ID once, including trains where data is missing.train_id_coordinates()excludes missing trains, and repeats train IDs if the source has multiple entries per train. The result will be the same length as the first dimension of an array fromndarray(), and tells you which train each entry belongs to.See also
xarray()returns a labelled array including these train IDs.
- select_trains(trains)
Select a subset of trains in this data as a new
KeyDataobject.Also available by slicing and indexing the KeyData object:
run[source, key][:10] # Select data for first 10 trains
- split_trains(parts=None, trains_per_part=None)
Split this data into chunks with a fraction of the trains each.
Either parts or trains_per_part must be specified.
This returns an iterator yielding new
KeyDataobjects. The parts will have similar sizes, e.g. splitting 11 trains withtrains_per_part=8will produce 5 & 6 trains, not 8 & 3. Selected trains count even if they are missing data, so different keys from the same run can be split into matching chunks.- Parameters:
parts (int) – How many parts to split the data into. If trains_per_part is also specified, this is a minimum, and it may make more parts. It may also make fewer if there are fewer trains in the data.
trains_per_part (int) – A maximum number of trains in each part. Parts will often have fewer trains than this.
New in version 1.7.
- as_single_value(rtol=1e-05, atol=0.0, reduce_by='median')
Retrieve a single reduced value if within tolerances.
The relative and absolute tolerances rtol and atol work the same way as in
numpy.allclose. The default relative tolerance is 1e-5 with no absolute tolerance. The data for this key is compared against a reduced value obtained by the method described in reduce_by.This may be a callable taking the key data, the string value of a global symbol in the numpy packge such as ‘median’ or ‘first’ to use the first value encountered. By default, ‘median’ is used.
If within tolerances, the reduced value is returned.
New in version 1.9.
- units
The units symbol for this data, e.g. ‘μJ’, or None if not found
- units_name
The units name for this data, e.g. ‘microjoule’, or None if not found
The run or file object (a DataCollection) also has methods to load
data by sources and keys. get_array(), get_dask_array() and
get_series() are directly equivalent to the options above, but other
methods offer extra capabilities.
- class extra_data.DataCollection
- get_array(source, key, extra_dims=None, roi=(), name=None)
Return a labelled array for a data field defined by source and key.
see
KeyData.xarray()for details.
- get_dask_array(source, key, labelled=False)
Get a Dask array for a data field defined by source and key.
see
KeyData.dask_array()for details.
- get_series(source, key)
Return a pandas Series for a 1D data field defined by source & key.
See
KeyData.series()for details.
- get_dataframe(fields=None, *, timestamps=False)
Return a pandas dataframe for given data fields.
df = run.get_dataframe(fields=[ ("*_XGM/*", "*.i[xy]Pos"), ("*_XGM/*", "*.photonFlux") ])
This links together multiple 1-dimensional datasets as columns in a table.
- Parameters:
See also
- pandas documentation
How to use the objects returned by
get_series()andget_dataframe()- Reading data to analyse in memory
Examples using xarray & pandas with EuXFEL data
- get_virtual_dataset(source, key, filename=None)
Create an HDF5 virtual dataset for a given source & key
A dataset looks like a multidimensional array, but the data is loaded on-demand when you access it. So it’s suitable as an interface to data which is too big to load entirely into memory.
This returns an h5py.Dataset object. This exists in a real file as a ‘virtual dataset’, a collection of links pointing to the data in real datasets. If filename is passed, the file is written at that path, overwriting if it already exists. Otherwise, it uses a new temp file.
To access the dataset from other worker processes, give them the name of the created file along with the path to the dataset inside it (accessible as
ds.name). They will need at least HDF5 1.10 to access the virtual dataset, and they must be on a system with access to the original data files, as the virtual dataset points to those.
- get_run_value(source, key)
Get a single value from the RUN section of data files.
RUN records each property of control devices as a snapshot at the beginning of the run. This includes properties which are not saved continuously in CONTROL data.
This method is intended for use with data from a single run. If you combine data from multiple runs, it will raise MultiRunError.
- Parameters:
New in version 1.6.
Getting data by train
Some kinds of data, e.g. from AGIPD, are too big to load a whole run into memory at once. In these cases, it’s convenient to load one train at a time.
If you want to do this for just one source & key with KeyData methods,
like this:
for tid, arr in run['SA1_XTD2_XGM/XGM/DOOCS:output', 'data.intensityTD'].trains():
...
- class extra_data.KeyData
- trains(keep_dims=False, include_empty=False)
Iterate through trains containing data for this key
Yields pairs of (train ID, array). Train axis is removed in case of single elements unless keep_dims is set. Skips trains where data is missing unless include_empty is set, returning None or zero-length array with keep_dims.
- train_from_id(tid, keep_dims=False)
Get data for the given train ID as a numpy array.
Returns (train ID, array)
- train_from_index(i, keep_dims=False)
Get data for a train by index (starting at 0) as a numpy array.
Returns (train ID, array)
To work with multiple modules of the same detector, see Multi-module detector data.
You can also get data by train for multiple sources and keys together from a run
or file object.
It’s always a good idea to select the data you’re interested in, either using
select(), or the devices= parameter. If you don’t,
they will read data for all sources in the run, which may be very slow.
- class extra_data.DataCollection
- trains(devices=None, train_range=None, *, require_all=False, flat_keys=False, keep_dims=False)
Iterate over all trains in the data and gather all sources.
run = Run('/path/to/my/run/r0123') for train_id, data in run.select("*/DET/*", "image.data").trains(): mod0 = data["FXE_DET_LPD1M-1/DET/0CH0:xtdf"]["image.data"]
- Parameters:
devices (dict or list, optional) – Filter data by sources and keys. Refer to
select()for how to use this.train_range (by_id or by_index object, optional) – Iterate over only selected trains, by train ID or by index. Refer to
select_trains()for how to use this.require_all (bool) – False (default) returns any data available for the requested trains. True skips trains which don’t have all the selected data; this only makes sense if you make a selection with devices or
select().flat_keys (bool) – False (default) returns nested dictionaries in each iteration indexed by source and then key. True returns a flat dictionary indexed by (source, key) tuples.
keep_dims (bool) – False (default) drops the first dimension when there is a single entry. True preserves this dimension.
- Yields:
tid (int) – The train ID of the returned train
data (dict) – The data for this train, keyed by device name
- train_from_id(train_id, devices=None, *, flat_keys=False, keep_dims=False)
Get train data for specified train ID.
- Parameters:
train_id (int) – The train ID
devices (dict or list, optional) – Filter data by sources and keys. Refer to
select()for how to use this.flat_keys (bool) – False (default) returns a nested dict indexed by source and then key. True returns a flat dictionary indexed by (source, key) tuples.
keep_dims (bool) – False (default) drops the first dimension when there is a single entry. True preserves this dimension.
- Returns:
tid (int) – The train ID of the returned train
data (dict) – The data for this train, keyed by device name
- Raises:
KeyError – if train_id is not found in the run.
- train_from_index(train_index, devices=None, *, flat_keys=False, keep_dims=False)
Get train data of the nth train in this data.
- Parameters:
train_index (int) – Index of the train in the file.
devices (dict or list, optional) – Filter data by sources and keys. Refer to
select()for how to use this.flat_keys (bool) – False (default) returns a nested dict indexed by source and then key. True returns a flat dictionary indexed by (source, key) tuples.
keep_dims (bool) – False (default) drops the first dimension when there is a single entry. True preserves this dimension.
- Returns:
tid (int) – The train ID of the returned train
data (dict) – The data for this train, keyed by device name
Selecting & combining data
These methods all return a new DataCollection object with the selected
data, so you use them like this:
sel = run.select("*/XGM/*")
# sel includes only XGM sources
# run still includes all the data
- class extra_data.DataCollection
- select(seln_or_source_glob, key_glob='*', require_all=False, require_any=False, *, warn_drop_trains_frac=1.0)
Select a subset of sources and keys from this data.
There are four possible ways to select data:
With two glob patterns (see below) for source and key names:
# Select data in the image group for any detector sources sel = run.select('*/DET/*', 'image.*')
With an iterable of source glob patterns, or (source, key) patterns:
# Select image.data and image.mask for any detector sources sel = run.select([('*/DET/*', 'image.data'), ('*/DET/*', 'image.mask')]) # Select & align undulator & XGM devices sel = run.select(['*XGM/*', 'MID_XTD1_UND/DOOCS/ENERGY'], require_all=True)
Data is included if it matches any of the pattern pairs.
With a dict of source names mapped to sets of key names (or empty sets to get all keys):
# Select image.data from one detector source, and all data from one XGM sel = run.select({'SPB_DET_AGIPD1M-1/DET/0CH0:xtdf': {'image.data'}, 'SA1_XTD2_XGM/XGM/DOOCS': set()})
Unlike the others, this option doesn’t allow glob patterns. It’s a more precise but less convenient option for code that knows exactly what sources and keys it needs.
With an existing DataCollection, SourceData or KeyData object:
# Select the same data contained in another DataCollection prev_run.select(sel)
The optional require_all and require_any arguments restrict the trains to those for which all or at least one selected sources and keys have at least one data entry. By default, all trains remain selected.
With require_all=True, a warning will be shown if there are no trains with all the required data. Setting warn_drop_trains_frac can show the same warning if there are a few remaining trains. This is a number 0-1 representing the fraction of trains dropped for one source (default 1).
Returns a new
DataCollectionobject for the selected data.Note
‘Glob’ patterns may be familiar from selecting files in a Unix shell.
*matches anything, so*/DET/*selects sources with “/DET/” anywhere in the name. There are several kinds of wildcard:*: anything?: any single character[xyz]: one character, “x”, “y” or “z”[0-9]: one digit character[!xyz]: one character, not x, y or z
Anything else in the pattern must match exactly. It’s case-sensitive, so “x” does not match “X”.
- AliasIndexer.select(seln_or_alias, key_glob='*', require_all=False, require_any=False)
Select a subset of sources and keys from this data using aliases.
This method is only accessible through the
DataCollection.aliasproperty.In contrast to
DataCollection.select(), only a subset of ways to select data via aliases is supported:With a source alias and literal key glob pattern:
# Select all pulse energy keys for an aliased XGM fast data. sel = run.alias.select('sa1-xgm', 'data.intensity*')
With an iterable of aliases and/or (source alias, key pattern) tuples:
# Select specific keys for an aliased XGM fast data. sel = run.alias.select([('sa1-xgm', 'data.intensitySa1TD'), ('sa1-xgm', 'data.intensitySa3TD')] # Select several aliases, may be both source and key aliases. sel = run.alias.select(['sa1-xgm', 'mono-hv'])
Data is included if it matches any of the aliases. Note that this method does not support glob patterns for the source alias.
With a dict of source aliases mapped to sets of key names (or empty sets to get all keys):
# Select image.data from an aliased AGIPD and all data # from an aliased XGM. sel = run.select({'agipd': {'image.data'}, 'sa1-xgm': set()})
The optional require_all and require_any arguments restrict the trains to those for which all or at least one selected sources and keys have at least one data entry. By default, all trains remain selected.
Returns a new
DataCollectionobject for the selected data.
- deselect(seln_or_source_glob, key_glob='*')
Select everything except the specified sources and keys.
This takes the same arguments as
select(), but the sources and keys you specify are dropped from the selection.Returns a new
DataCollectionobject for the remaining data.
- AliasIndexer.deselect(seln_or_alias, key_glob='*')
Select everything except the specified sources and keys using aliases.
This method is only accessible through the
DataCollection.aliasproperty.This takes the same arguments as
select(), but the sources and keys you specify are dropped from the selection.Returns a new
DataCollectionobject for the remaining data.
- select_trains(train_range)
Select a subset of trains from this data.
Slice trains by position within this data:
sel = run.select_trains(np.s_[:5])
Or select trains by train ID, with a slice or a list:
from extra_data import by_id sel1 = run.select_trains(by_id[142844490 : 142844495]) sel2 = run.select_trains(by_id[[142844490, 142844493, 142844494]])
Returns a new
DataCollectionobject for the selected trains.- Raises:
ValueError – If given train IDs do not overlap with the trains in this data.
- split_trains(parts=None, trains_per_part=None)
Split this data into chunks with a fraction of the trains each.
Either parts or trains_per_part must be specified.
This returns an iterator yielding new
DataCollectionobjects. The parts will have similar sizes, e.g. splitting 11 trains withtrains_per_part=8will produce 5 & 6 trains, not 8 & 3.- Parameters:
parts (int) – How many parts to split the data into. If trains_per_part is also specified, this is a minimum, and it may make more parts. It may also make fewer if there are fewer trains in the data.
trains_per_part (int) – A maximum number of trains in each part. Parts will often have fewer trains than this.
New in version 1.7.
- union(*others)
Join the data in this collection with one or more others.
This can be used to join multiple sources for the same trains, or to extend the same sources with data for further trains. The order of the datasets doesn’t matter. Any aliases defined on the collections are combined as well unless their values conflict.
Note that the trains for each source are unioned as well, such that
run.train_ids == run[src].train_ids.Returns a new
DataCollectionobject.
Writing selected data
- class extra_data.DataCollection
- write(filename)
Write the selected data to a new HDF5 file
You can choose a subset of the data using methods like
select()andselect_trains(), then use this write it to a new, smaller file.The target filename will be overwritten if it already exists.
- write_virtual(filename)
Write an HDF5 file with virtual datasets for the selected data.
This doesn’t copy the data, but each virtual dataset provides a view of data spanning multiple sequence files, which can be accessed as if it had been copied into one big file.
This is not the same as building virtual datasets to combine multi-module detector data. See Multi-module detector data for that.
Creating and reading virtual datasets requires HDF5 version 1.10.
The target filename will be overwritten if it already exists.
Missing data
What happens if some data was not recorded for a given train?
Control data is duplicated for each train until it changes. If the device cannot send changes, the last values will be recorded for each subsequent train until it sends changes again. There is no general way to distinguish this scenario from values which genuinely aren’t changing.
Parts of instrument data may be missing from the file. These will also be
missing from the data returned by extra_data:
The train-oriented methods
trains(),train_from_id(), andtrain_from_index()give you dictionaries keyed by source and key name. Sources and keys are only included if they have data for that train.get_array(), andget_series()skip over trains which are missing data. The indexes on the returned DataArray or Series objects link the returned data to train IDs. Further operations with xarray or pandas may drop misaligned data or introduce fill values.get_dataframe()includes rows for which any column has data. Where some but not all columns have data, the missing values are filled withNaNby pandas’ missing data handling.
Missing data does not necessarily mean that something has gone wrong: some devices send data at less than 10 Hz (the train rate), so they always have gaps between updates.
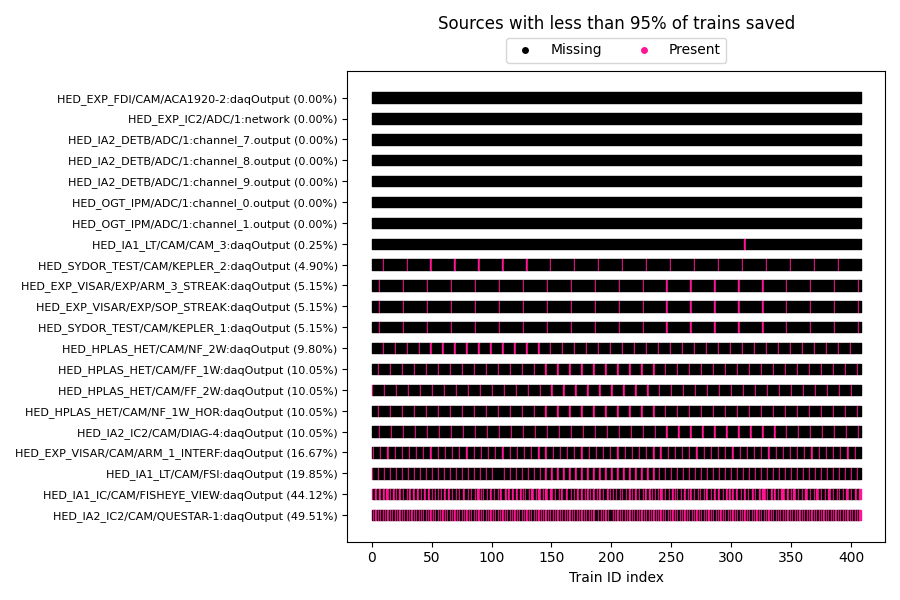
You can use DataCollection.plot_missing_data() to quickly visualize
sources that have missing data:
- class extra_data.DataCollection
- plot_missing_data(min_saved_pct=95, expand_instrument=False)
Plot sources that have missing data for some trains.
Example output:
Data problems
If you encounter problems accessing data with extra_data, there may be
problems with the data files themselves. Use the extra-data-validate
command to check for this (see Checking data files).
Here are some problems we’ve seen, and possible solutions or workarounds:
Indexes point to data beyond the end of datasets: this has previously been caused by bugs in the detector calibration pipeline. If you see this in calibrated data (in the
proc/folder), ask for the relevant runs to be re-calibrated.Train IDs are not strictly increasing: issues with the timing system when the data is recorded can create an occasional train ID which is completely out of sequence. Usually it seems to be possible to ignore this and use the remaining data, but if you have any issues, please let us know.
In one case, a train ID had the maximum possible value (264 - 1), causing
info()to fail. You can select everything except this train usingselect_trains():from extra_data import by_id sel = run.select_trains(by_id[:2**64-1])
If you’re having problems with extra_data, you can also try searching previously reported issues to see if anyone has encountered similar symptoms.
Cached run data maps
When you open a run in extra_data, it needs to know what data is in each file. Each file has metadata describing its contents, but reading this from every file is slow, especially on GPFS. extra_data therefore tries to cache this information the first time a run is opened, and reuse it when opening that run again.
This should happen automatically, without the user needing to know about it. You only need these details if you think caching may be causing problems.
Caching is triggered when you use
RunDirectory()oropen_run().There are two possible locations for the cached data map:
In the run directory:
(run dir)/karabo_data_map.json.In the proposal scratch directory:
(proposal dir)/scratch/.karabo_data_maps/raw_r0032.json. This will normally be the one used on Maxwell, as users can’t write to the run directory.
The format is a JSON array, with an object for each file in the run.
This holds the list of train IDs in the file, and the lists of control and instrument sources.
It also stores the file size and last modified time of each data file, to check if the file has changed since the cache was created. If either of these attributes doesn’t match, extra_data ignores the cached information and reads the metadata from the HDF5 file.
If any file in the run wasn’t listed in the data map, or its entry was outdated, a new data map is written automatically. It tries the same two locations described above, but it will continue without error if it can’t write to either.
JSON was chosen as it can be easily inspected manually, and it’s reasonably efficient to load the entire file.
Issues reading archived data
Files at European XFEL storage migrate over time from GPFS (designed for fast access) to PNFS (designed for archiving). The data on PNFS is usually always available for reading. But sometimes, this may require staging from the tape to disk. If there is a staging queue, the operation can take an indefinitely long time (days or even weeks) and any IO operations will be blocked for this time.
To determine the files which require staging or are lost, use the script:
extra-data-locality <run directory>
It returns a list of files which are currently located only on slow media for some reasons and, separately, any which have been lost.
If the files are not essential for analysis, then they can be filtered out using
filter lc_ondisk() from extra_data.locality:
from extra_data.locality import lc_ondisk
run = open_run(proposal=700000, run=1, file_filter=lc_ondisk)
file_filter must be a callable which takes a list as a single argument and
returns filtered list.
Note: Reading the file locality on PNFS is an expensive operation. Use it only as a last resort.
If you find any files which are located only on tape or unavailable, please let know to ITDM. If you need these files for analysis mentioned that explicitly.
‘Suspect’ train IDs
In some cases (especially with AGIPD data), some train IDs appear to be recorded
incorrectly, breaking the normal assumption that train IDs are in increasing
order. EXtra-data will exclude these trains by default, but you can try to
access them by passing inc_suspect_trains=True when opening a file
or run. Some features may not work correctly if you do this.
In newer files (format version 1.0 or above), trains are considered suspect
where their INDEX/flag entry is 0. This indicates that the DAQ received the train ID
from a device before it received it from a time server. This appears to be a reliable
indicator of erroneous train IDs.
In older files without INDEX/flag, EXtra-data tries to guess which trains
are suspect. The good trains should make an increasing sequence, and it tries to
exclude as few trains as possible to achieve this. If something goes wrong with
this guessing, try using inc_suspect_trains=True to avoid it.
Please let us know (da-support@xfel.eu) if you need to do this.